
I am currently a Ph.D. candidate at the University of Twente in the Netherlands, under the supervision of Dr. Doina Bucur and Prof. Maurice van Keulen. Before joining UT, I was a Machine Learning Engineer at Baidu, Inc.
My research interests lie in the fields of network data science and machine learning. I used to work on brain networks, but now I am shifting to microbial and ecological networks. I mainly specialize in network structure inference, network classification, dynamic/multimodal network analysis, causal inference, and further practical applications based on networks.
Warning
Problem: The current name of your GitHub Pages repository ("Solution: Please consider renaming the repository to "
http://".
However, if the current repository name is intended, you can ignore this message by removing "{% include widgets/debug_repo_name.html %}" in index.html.
Action required
Problem: The current root path of this site is "baseurl ("_config.yml.
Solution: Please set the
baseurl in _config.yml to "Education
-
 University of TwenteAug. 2023 - present
University of TwenteAug. 2023 - present -
 Lanzhou UniversityM.E. in Computer ScienceSep. 2019 - Jul. 2022
Lanzhou UniversityM.E. in Computer ScienceSep. 2019 - Jul. 2022 -
 Taiyuan University of TechenologyB.E. in AutomationSep. 2014 - Jul. 2018
Taiyuan University of TechenologyB.E. in AutomationSep. 2014 - Jul. 2018
Experience
-
 Baidu, Inc.Machine Learning EngineerJul. 2022 - Jul. 2023
Baidu, Inc.Machine Learning EngineerJul. 2022 - Jul. 2023 -
 Inspur Co., Ltd.Software Development EngineerJul. 2018 - Apr. 2019
Inspur Co., Ltd.Software Development EngineerJul. 2018 - Apr. 2019
Honors & Awards (Recent)
-
Honored Graduate of Lanzhou University and Gansu Province2022
-
Graduate Student of the Year2021
-
National Scholarship2021
Selected Publications (view all )
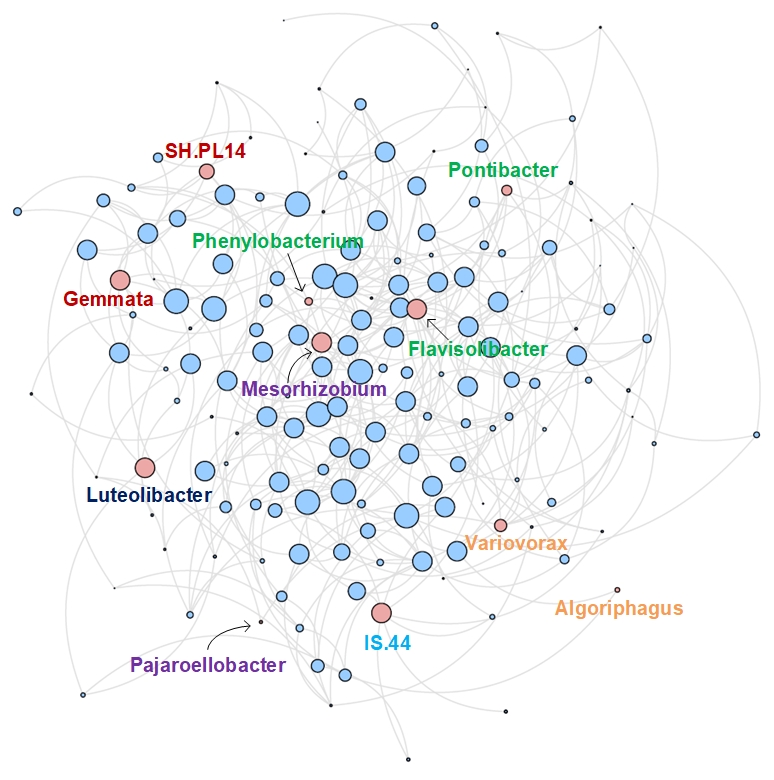
gFlora: a topology-aware method to discover functional co-response groups in soil microbial communities
Nan Chen, Merlijn Schram, Doina Bucur
IEEE Transactions on Computational Biology and Bioinformatics, 2024 Spotlight
Microorganisms such as bacteria perform critical functions in the soil ecosystem: they mediate essential carbon, nitrogen, and nutrient cycling processes in soils. To manage the health and functions of soils, it is important to understand which soil functions are related the most to which microbial taxa—but this taxon-to-function link is difficult to discover because of the size and complexity of the soil ecosystem. A feasible solution is to discover functional links at the level of groups instead of individuals, using observational data of both taxa abundance and soil function indicators. We thus aim to learn the functional co-response group: a group of taxa whose co-response effect (the representative characteristic of the whole functional group) co-responds (associates well statistically) to a functional variable. Unlike the existing method, we model the soil microbial community as an ecological co-occurrence network with the taxa as nodes (weighted by their abundance) and their relationships (a combination from both spatial and functional ecological aspects) as edges (weighted by the strength of the relationships). Then, we design a method called gFlora which notably uses graph convolution over this co-occurrence network to compute the co-response effect of the group, such that the network topology is also considered in the discovery process. We evaluate gFlora on four real-world soil microbiome datasets (bacteria and nematodes combined with two soil functions: nitrogen mineralization and crop yield). gFlora outperforms the competing method on all evaluation metrics, and it discovers new functional evidence for taxa which were so far under-studied. We show that the graph convolution is crucial to taxa with relatively low abundance (thus removing the bias towards taxa with higher abundance), and the discovered bacteria of different genera are distributed in the co-occurrence network but remain tightly connected among themselves, demonstrating that topologically they fill different but collaborative functional roles in the ecological community.
gFlora: a topology-aware method to discover functional co-response groups in soil microbial communities
Nan Chen, Merlijn Schram, Doina Bucur
IEEE Transactions on Computational Biology and Bioinformatics, 2024 Spotlight
Microorganisms such as bacteria perform critical functions in the soil ecosystem: they mediate essential carbon, nitrogen, and nutrient cycling processes in soils. To manage the health and functions of soils, it is important to understand which soil functions are related the most to which microbial taxa—but this taxon-to-function link is difficult to discover because of the size and complexity of the soil ecosystem. A feasible solution is to discover functional links at the level of groups instead of individuals, using observational data of both taxa abundance and soil function indicators. We thus aim to learn the functional co-response group: a group of taxa whose co-response effect (the representative characteristic of the whole functional group) co-responds (associates well statistically) to a functional variable. Unlike the existing method, we model the soil microbial community as an ecological co-occurrence network with the taxa as nodes (weighted by their abundance) and their relationships (a combination from both spatial and functional ecological aspects) as edges (weighted by the strength of the relationships). Then, we design a method called gFlora which notably uses graph convolution over this co-occurrence network to compute the co-response effect of the group, such that the network topology is also considered in the discovery process. We evaluate gFlora on four real-world soil microbiome datasets (bacteria and nematodes combined with two soil functions: nitrogen mineralization and crop yield). gFlora outperforms the competing method on all evaluation metrics, and it discovers new functional evidence for taxa which were so far under-studied. We show that the graph convolution is crucial to taxa with relatively low abundance (thus removing the bias towards taxa with higher abundance), and the discovered bacteria of different genera are distributed in the co-occurrence network but remain tightly connected among themselves, demonstrating that topologically they fill different but collaborative functional roles in the ecological community.
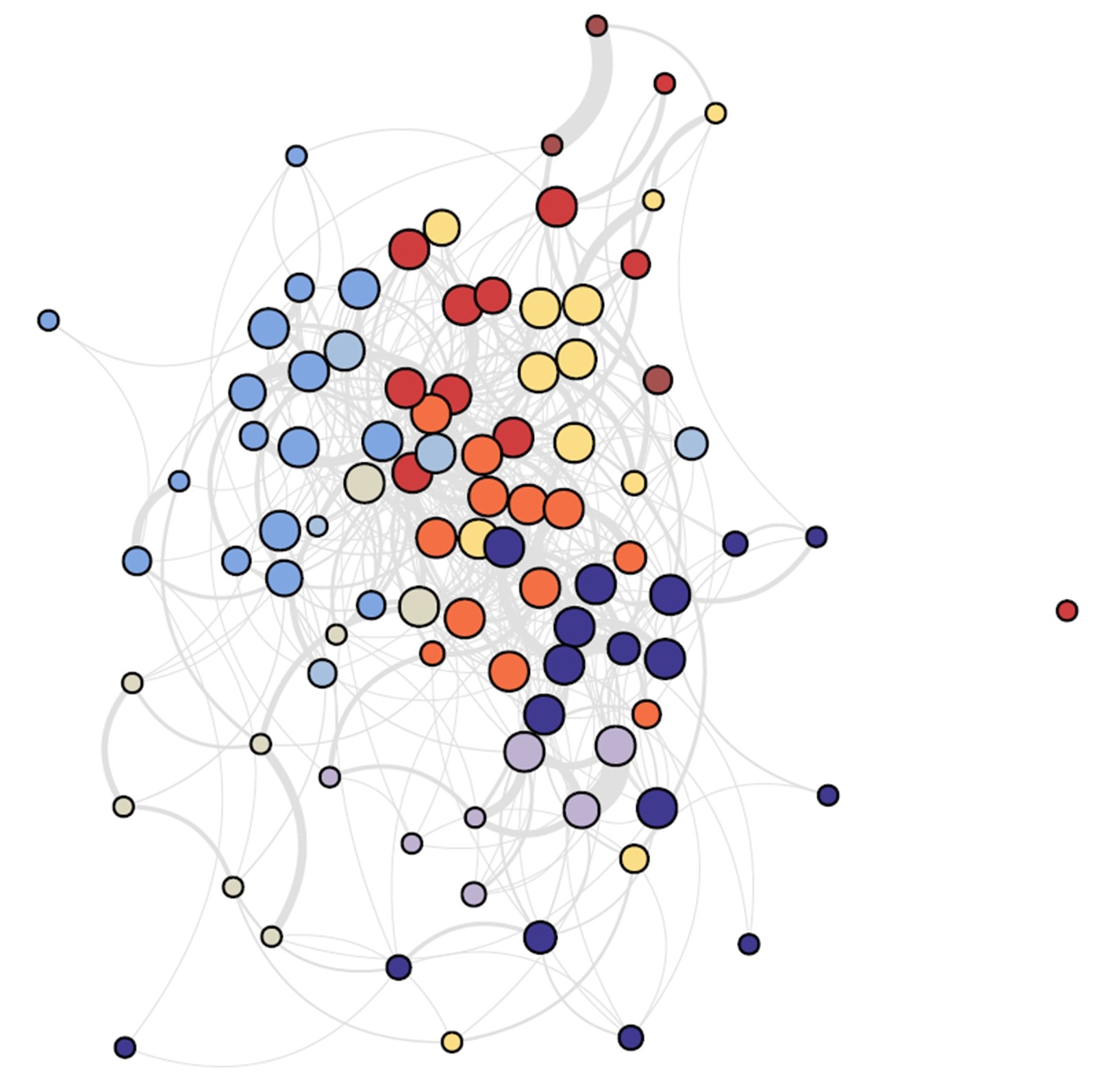
EleMi: A Robust Method to Infer Soil Ecological Networks with Better Community Structure
Nan Chen, Doina Bucur
International Conference on Complex Networks (CompleNet), 2024
Soil ecological networks enable us to better understand the complex interactions among a great number of organisms in soil. Soil communities are biotic groups with similar environmental and resource preferences. Community detection thus provides insights into the mechanisms of the soil ecosystem. Therefore, inferring ecological networks with clear community structure is essential for investigating the soil ecosystem. We propose Elastic net regularized Multi-regression (EleMi), a new method to infer soil ecological networks. To better find the community structure, EleMi does not infer pairwise interactions, but considers all organisms simultaneously. Specifically, it regresses the abundance of all other taxa to one taxon (with shared parameters across soil samples) and employs Elastic net to avoid over-sparsity and stochasticity. The results on both synthetic and real biotic data show that EleMi is more robust and can infer ecological networks with clearer community structure.
EleMi: A Robust Method to Infer Soil Ecological Networks with Better Community Structure
Nan Chen, Doina Bucur
International Conference on Complex Networks (CompleNet), 2024
Soil ecological networks enable us to better understand the complex interactions among a great number of organisms in soil. Soil communities are biotic groups with similar environmental and resource preferences. Community detection thus provides insights into the mechanisms of the soil ecosystem. Therefore, inferring ecological networks with clear community structure is essential for investigating the soil ecosystem. We propose Elastic net regularized Multi-regression (EleMi), a new method to infer soil ecological networks. To better find the community structure, EleMi does not infer pairwise interactions, but considers all organisms simultaneously. Specifically, it regresses the abundance of all other taxa to one taxon (with shared parameters across soil samples) and employs Elastic net to avoid over-sparsity and stochasticity. The results on both synthetic and real biotic data show that EleMi is more robust and can infer ecological networks with clearer community structure.